Gene Expression Analysis via GeneNominator™ to Find Biological Pathways
Helping to Bring Improved and Novel Therapies to the Right Patient Population Faster
GeneNominator™ is a bioinformatics tool that couples drug sensitivity data with gene expression data. It uses large datasets of gene expression to identify interconnections between sensitivity or resistance to your anti-cancer drug and gene transcription. Markers for your compounds that are associated with specific diseases will support the patient stratification for clinical trials.
Biological Markers of Protein Interactions Support the Development of Precision Medicine
Your anti-cancer drug candidate may act on a chain of biological changes. Features of the bioinformatics analysis that support the pathway analysis are:
Identification of Downstream Effects in Signalling Pathways
Gene expression analysis reveals correlations between drug sensitivity and elevated or decreased expression levels without prior knowledge on the affected biological processes. The comprehensive analysis includes mRNA data of more than 19,000 genes and can therefore find unexpected relationships. To support decision-making on next steps, we also perform the analysis on smaller subsets of genes, such as cancer genes and clinically actionable genes. Protein-protein interaction network plots are analysed for genes that are known to physically interact — for example, genes that are part of the same pathway.
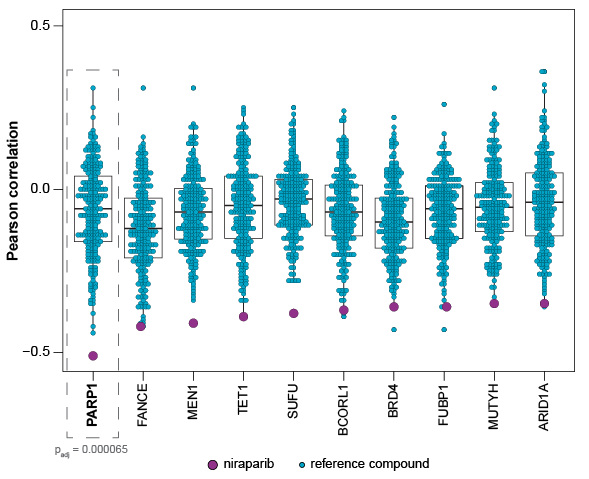
Your Compound’s Cancer Gene Expression Signature Is Compared to 248 Therapeutics
Unique biological markers for your compound are found by mapping the top-scoring genes against a collection of anti-cancer agents that represent a wide variety of mechanisms (e.g., PARP inhibitors, kinase inhibitors, epigenetic modulators). You can directly see that your compound has a distinct profile from competitive therapeutics.
References
Uitdehaag et al. (2019) Combined cellular and biochemical profiling to identify predictive drug response biomarkers for kinase inhibitors approved for clinical use between 2013 and 2017. Molecular Cancer Therapeutics, 18 (2):470-481.