Comparative Analysis via OncolinesProfiler™ to Identify Targets and Differentiators
Helping to Bring Improved and Novel Therapies to the Right Patient Population Faster
Comparative analysis is used to find or confirm the target of your compound. The compound may be of a unique scaffold or may have emerged from phenotypical screens, in which case the biochemical target may be speculative. The intention of repurposing drugs may also trigger the need for target identification or confirmation. Clustering analysis of a broad range of anti-cancer therapeutics, using the relative potency of compounds in proliferation assays, can reveal similarities or differences in the biochemical mode of action.
Mapping Your Compound in the Competitive Field
Clustering analysis via OncolinesProfiler™ compares your compound’s IC50 fingerprint against the cellular activity of 248 anti-cancer therapeutics. The latter include standards of care, clinical stage candidates and experimental therapeutics. The anti-cancer drug list covers different modalities and a wide variety of targets that are clinically applied or under investigation. Modalities include small molecule inhibitors, monoclonal antibodies, antibody-drug conjugates and degraders. Examples of targets are tubulin, proteasome, topoisomerase, ubiquitin pathway, bromodomain, kinases, Wnt pathway, RNA synthesis, HDAC, EZH2 and anti-apoptotic mechanisms.
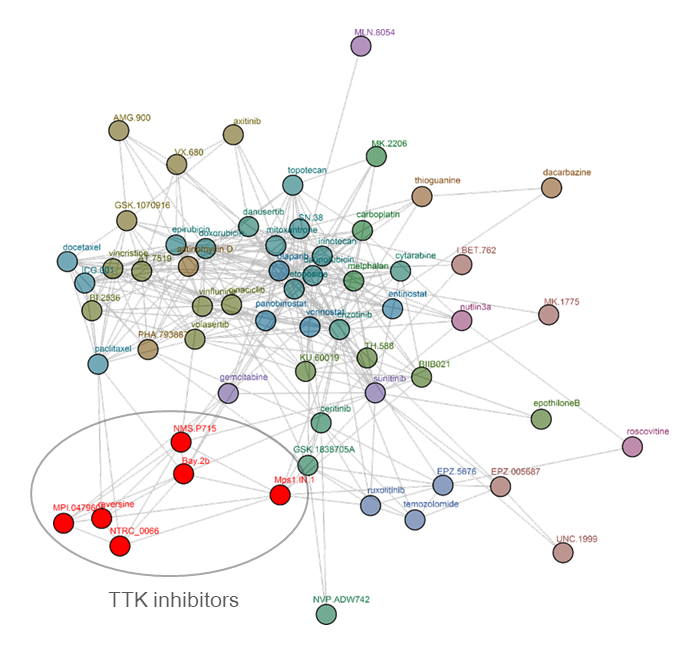
Clustering tree for reversine. Reversine was originally identified in a phenotypic screen for molecules that can promote dedifferentiation of myoblasts into multipotent progenitor-like cells (Chen et al., 2007), which was attributed to its ability to inhibit Aurora B (d’Alise et al., 2008). Later, it was shown that reversine can inhibit TTK (Santaguida et al., 2008). The comparative analysis shows that reversine targets TTK (Libouban et al., 2017). Other TTK inhibitors are pictured in red.
Best-in-Class and First-in-Class Inhibitors
The target for best-in-class agents is confirmed when your compound shares a cluster with the relevant targeted therapeutics. For instance, it may fall within the cluster of selective PI3K inhibitors instead of pan-PI3K inhibitors. Its best-in-class properties emerge from improved potency, increased tissue selectivity, or selectivity for a genomic biomarker.
When you are working on a novel mechanism, your compound will be unique and located separately from all other therapeutics in the dataset. The exact targeting may be found by, for instance, knockout experiments and zooming in on specific mechanisms via protein expression analysis.
References
Kooijman et al. (2022) Comparative kinase and cancer cell panel profiling of kinase inhibitors approved for clinical use form 2018 to 2020. Frontiers in Oncology, section Cancer Molecular Targets and Therapeutics, 12:953013.
Uitdehaag et al. (2016) Cell panel profiling reveals conserved therapeutic clusters and differentiates the mechanism of action of different PI3K/mTOR, Aurora kinase and EZH2 inhibitors. Molecular Cancer Therapeutics, 15 (12):3097-3109.
Chen et al. (2007) Reversine increases the plasticity of lineage-committed mammalian cells. PNAS, 104 (25):10482-10487.
d’Alise et al. (2008) Reversine, a novel Aurora kinases inhibitor, inhibits colony formation of human acute myeloid leukemia cells. Molecular Cancer Therapeutics, 7 (5):1140-1149.
Santaguida et al. (2008) Dissecting the role of MPS1 in chromosome biorientation and the spindle checkpoint through the small molecule inhibitor reversine. Journal of Cell Biology, 190 (1):73-87.
Libouban et al. (2017) Stable aneuploid tumors cells are more sensitive to TTK inhibition than chromosomally unstable cell lines. Oncotarget, 8 (24):38309-38325.