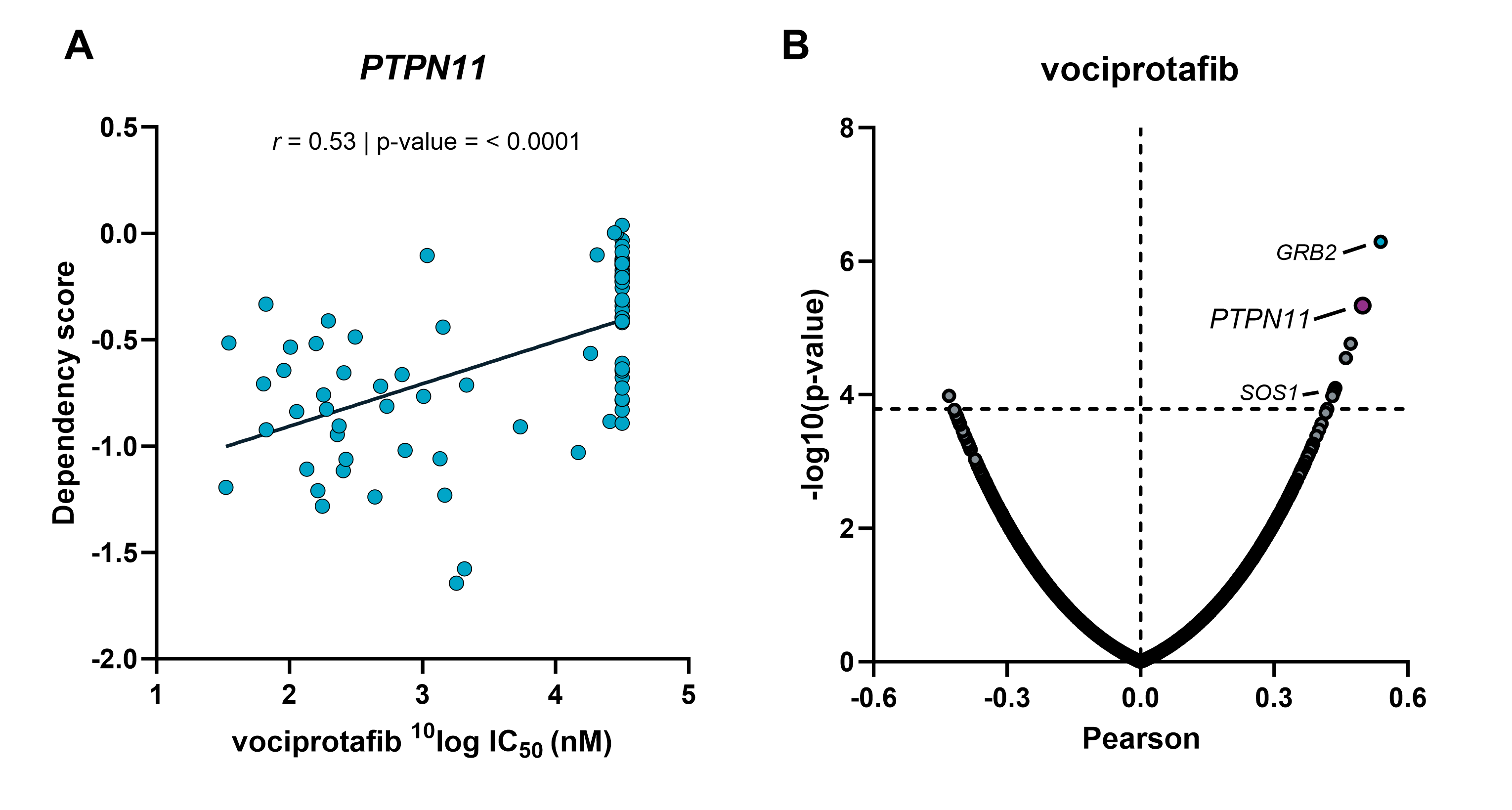
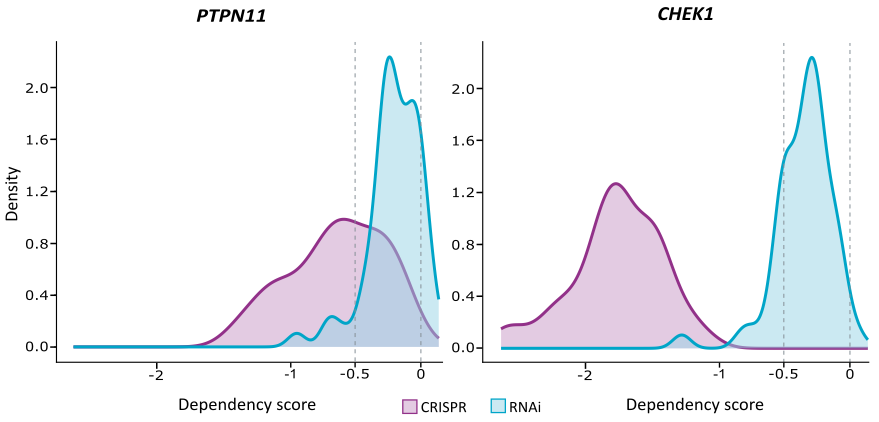
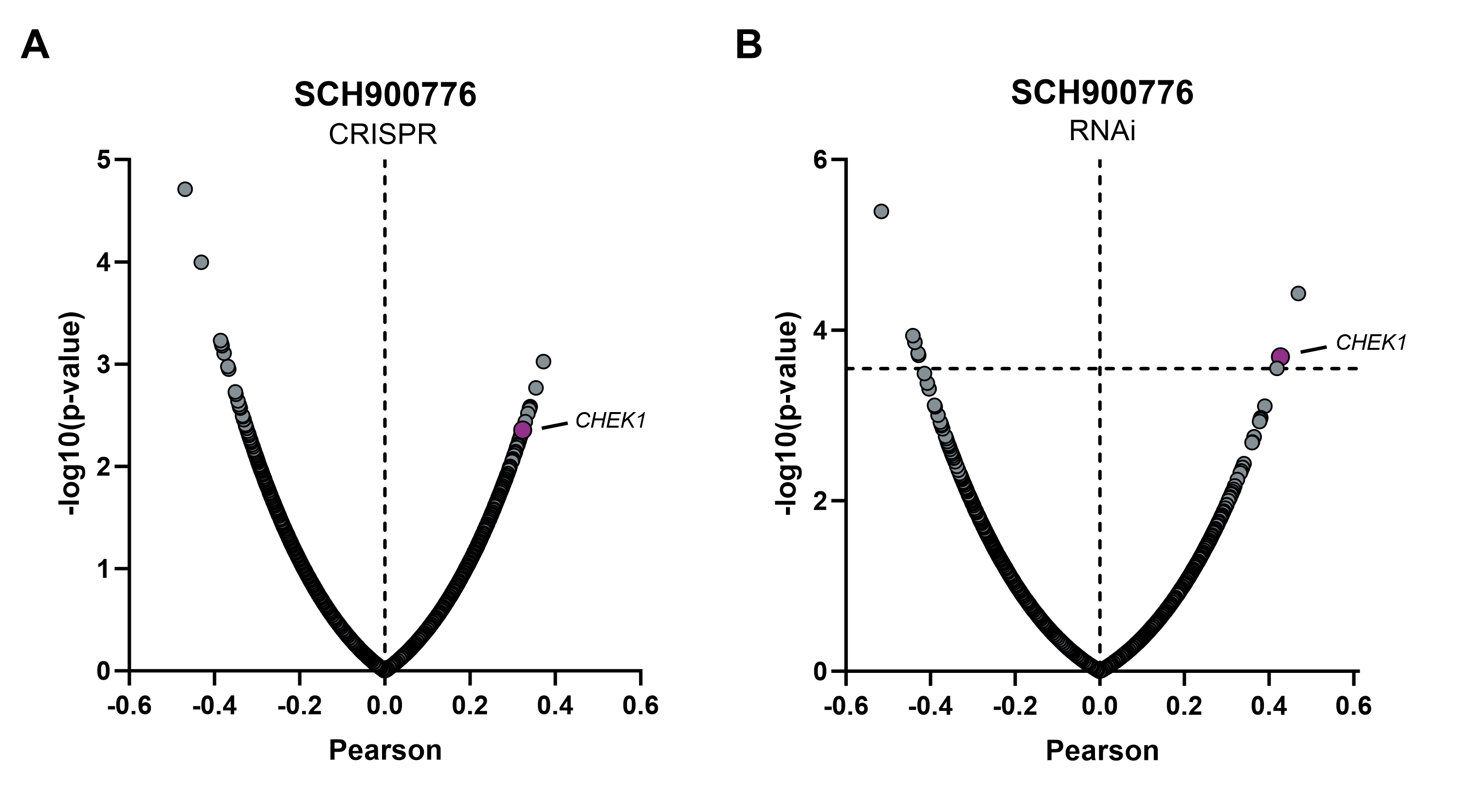
Target validation by combining Oncolines® profiling and gene dependency screens
References
1. Meyers et al. (2017) Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nature Genetics 49, 1779-1784.
2. Dempster et al. (2019) Extracting biological insights from the project Achilles genome-scale CRISPR screens in cancer cell lines. BioRxiv, 720243.
3. McFarland et al. (2018) Improved estimation of cancer dependencies from large-scale RNAi screens using model-based normalization and data integration. Nature Communications 9, 4610.
4. Dempster et al. (2021) Chronos: a cell population dynamics model of CRISPR experiments that improves inference of gene fitness effects. Genome Biology 22, 343
5. Pacini et al. (2021) Integrated cross-study datasets of genetic dependencies in cancer. Nature Communications 12, 1661.