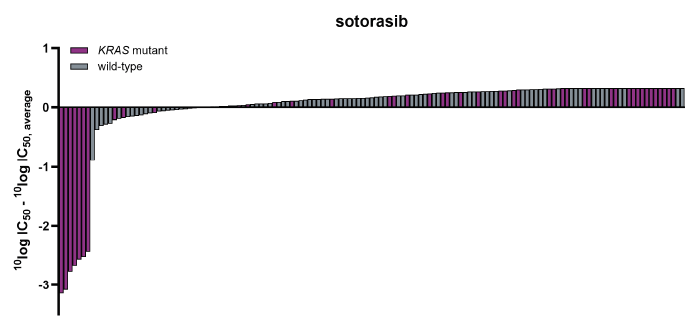
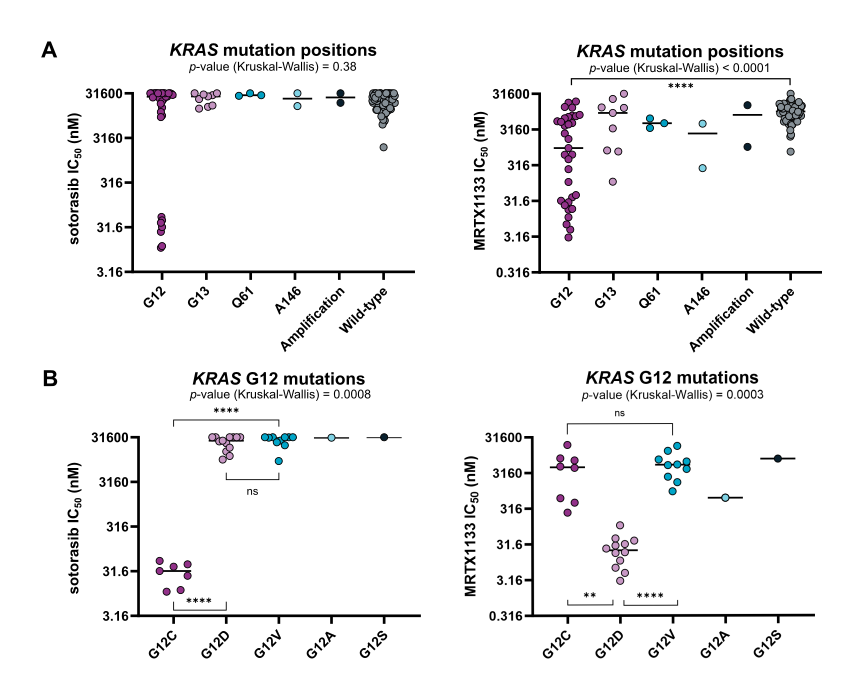
Stepwise, in-depth cancer gene mutation analysis reveals cellular targeting of KRAS mutant-selective inhibitors
References
1. Canon et al. (2019) The clinical KRAS G12C inhibitor AMG 510 drives anti-tumour immunity. Nature 575, 217–223.
2. Wang et al. (2022) Identification of MRTX1133, a noncovalent, potent, and selective KRASG12D inhibitor. J Med Chem 65, 3123–3133.