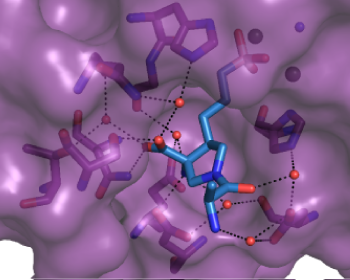
Molecular Interactions for In Vitro Proof-of-Concept Studies
Biochemical Characterisation of Inhibitor-Target Interaction for Precision Medicine
Selection of the best drug candidate for your target requires an understanding of the binding characteristics of the compounds to the target. Several biochemical techniques can be applied to select the compound with the best characteristics for further analysis.
Relating Drug Activity to Binding Characteristics via Biochemical Assays
The design of novel compounds with improved properties can be facilitated using a variety of assays. A typical workflow consists of 1. Protein expression and purification; 2. Development of an activity assay and selection of compounds; 3. Profiling compounds via SPR assay; 4. Thermal stability assay; 5. Crystal structure determination of the protein in the presence of promising inhibitors.
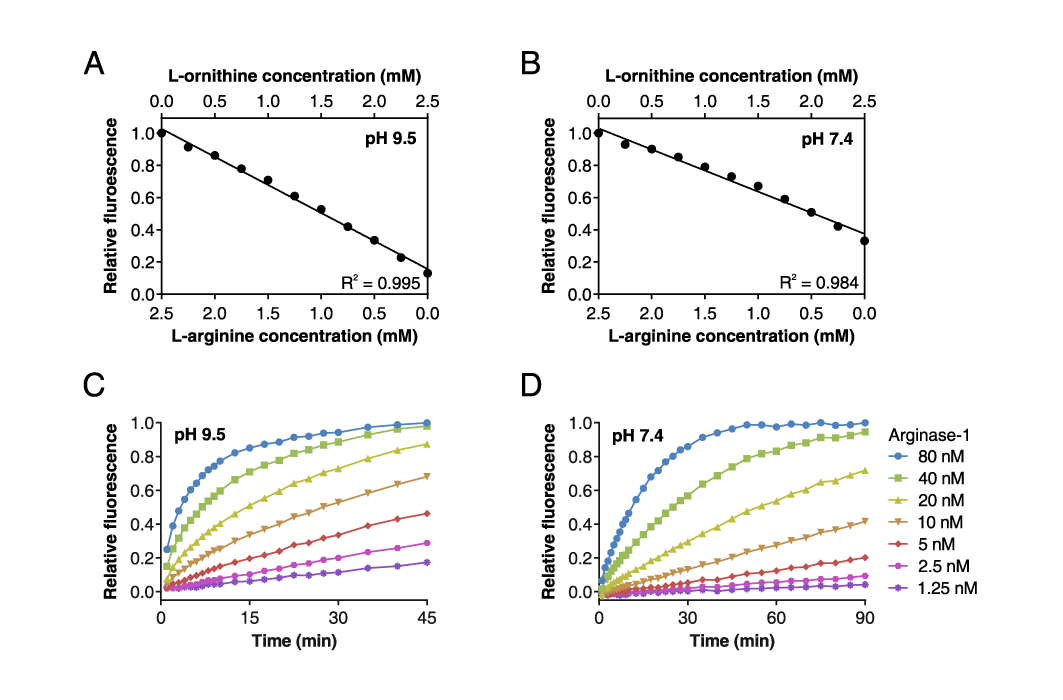
Fluorescence-based Arginase-1 activity assay
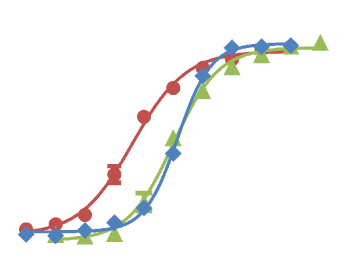
(A) Combined L-arginine and L-ornithine calibration curve at pH 9.5 and (B) pH 7.4, demonstrating that the fluorescent signal is linearly proportional to the L-arginine and L-ornithine concentrations within the measuring range of the assay.
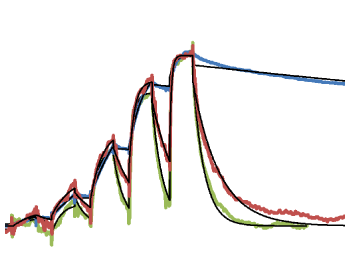
(C) Time-course of Arginase-1 activity at different enzyme concentrations at pH 9.5 and (D) at pH 7.4 measured in real time. (Adapted from Grobben et al., 2020).
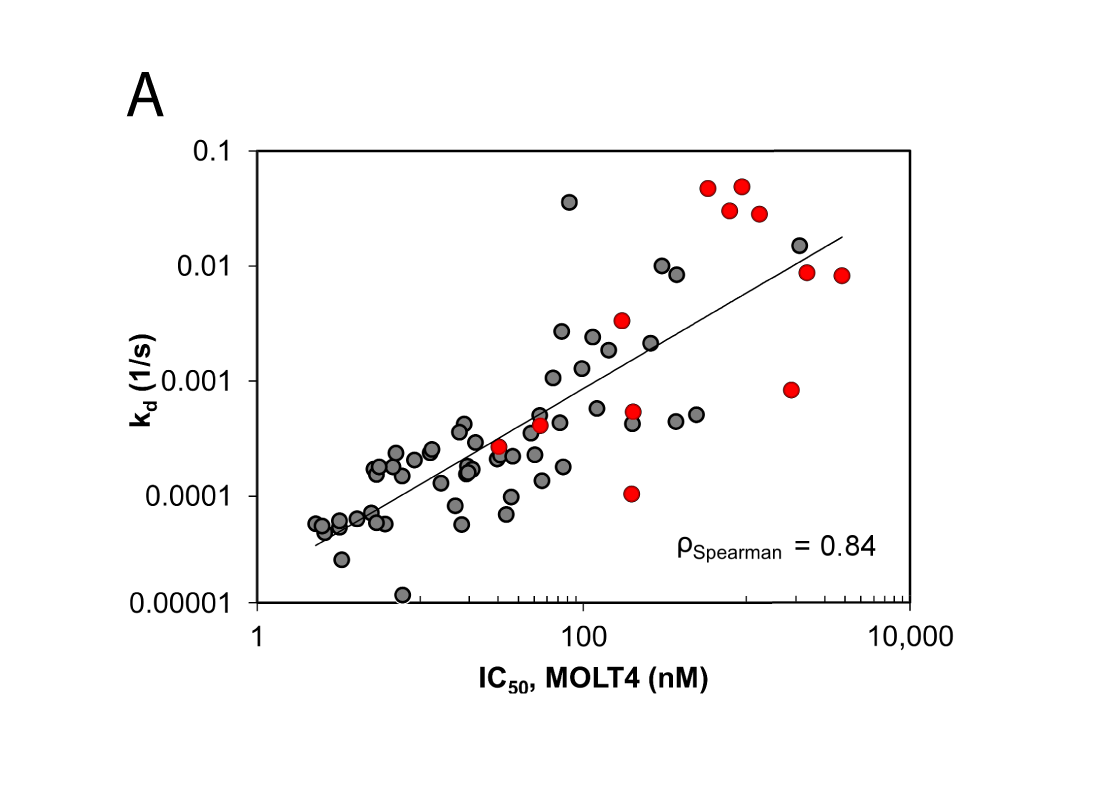
Deconvolution of Binding Mode, Site, and Mechanism
Our protein and biophysical scientists are in close contact with the sponsor to determine the most suitable assay format for the study and to share scientific insights. Oncolines also has expertise in cancer biology services. The biochemical and cell-based assay technologies provide a comprehensive portfolio for in vitro characterisation of your lead candidates.
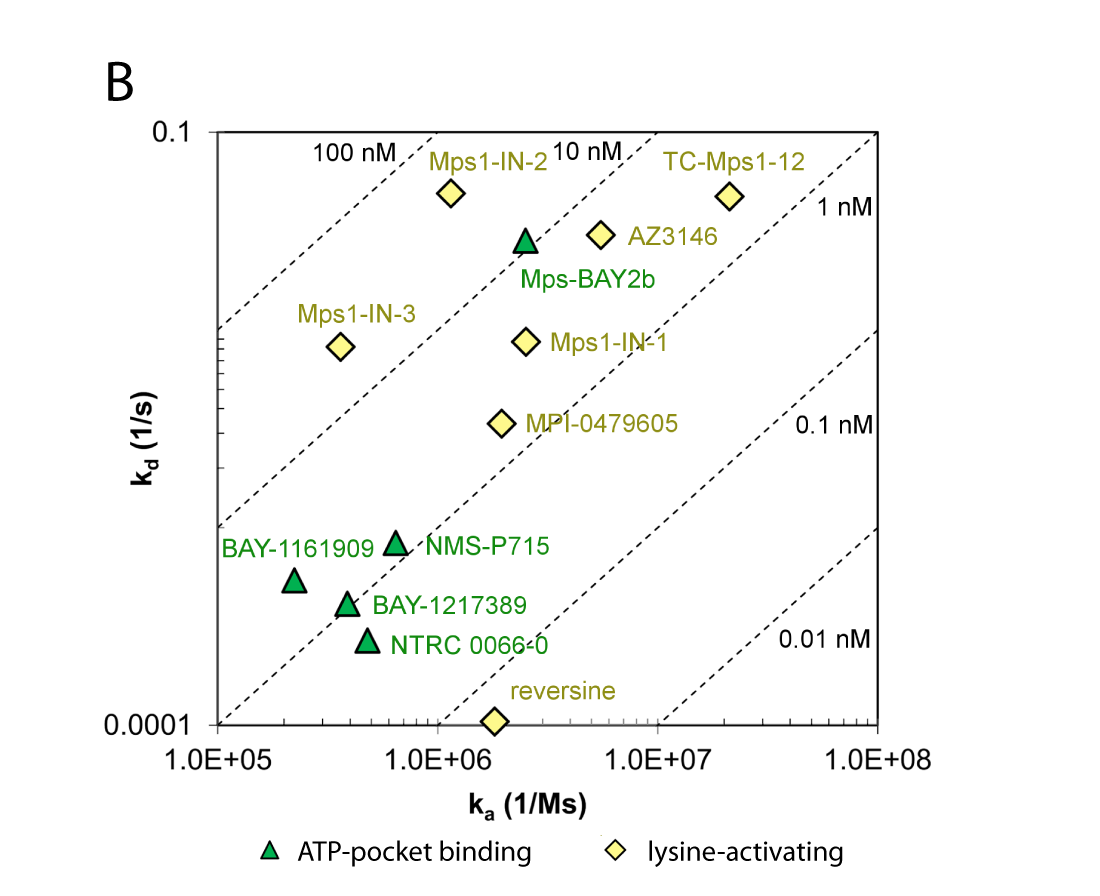
(B) Comparison of association and dissociation rates for reference inhibitors. Diagonals represent equipotency lines with pertinent KD values indicated within the frame. The ATP-pocket binding (green, triangles) and lysine-targeting inhibitors (yellow, lozenge) can be distinguished as separate groups.
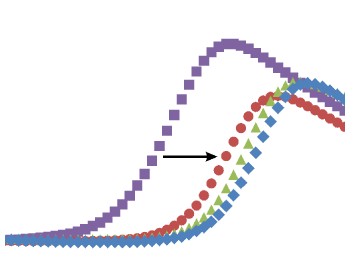
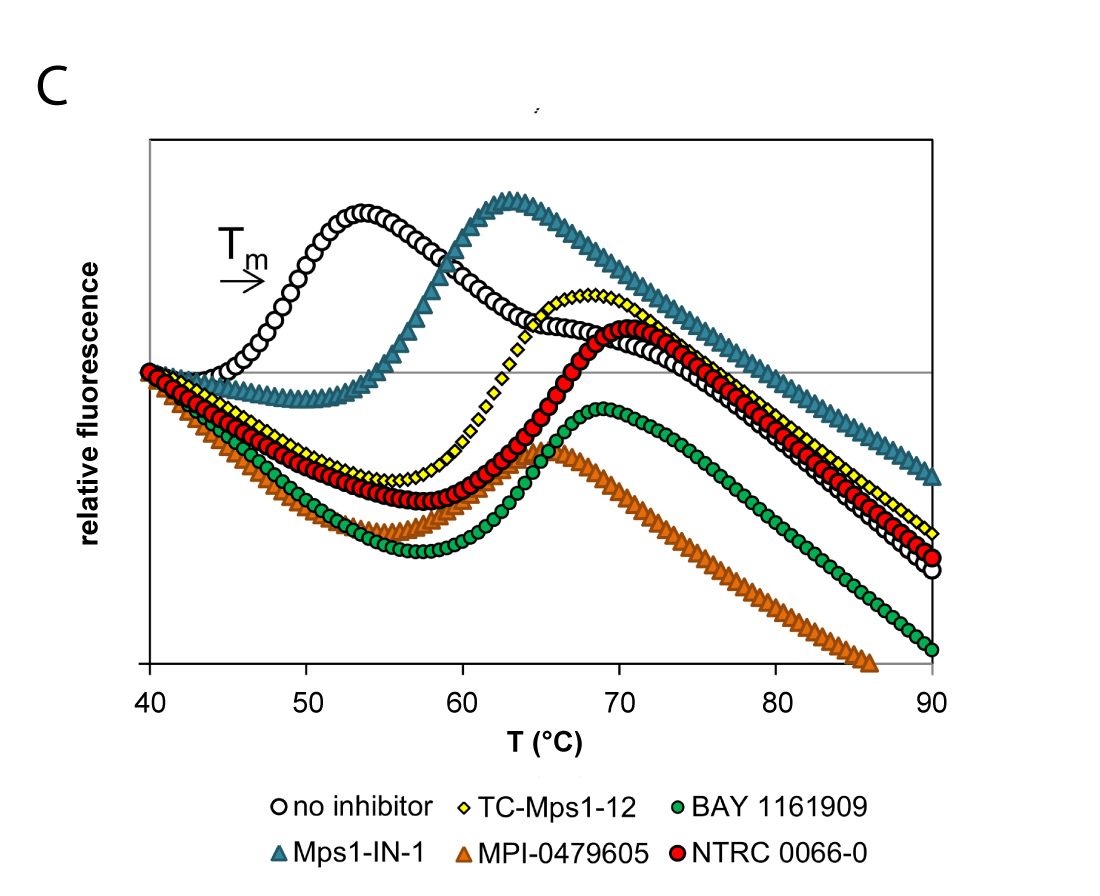
(C) Thermal unfolding curves of the TTK kinase domain in the presence of TTK inhibitors. Relatively fast dissociating compounds shift, unfolding to a lesser extent than slowly dissociating compounds. This indicates that the binding of TTK inhibitors with long target residence times induces a stabilizing conformational shift (Uitdehaag et al., 2017).
In Vitro Studies Enhance Discovery of Drugs for Oncology Precision Medicine
It is our mission to help you find a mechanistic hypothesis before entering the clinic. We have generated science-based technologies that help you to identify the mechanism of preclinical and clinical drug candidates. Our technologies provide in vitro proof of concept in the preparation of in vivo studies and clinical trials.
Deliverables You Can Count On
We deliver reproducible results that are adapted to your specific needs with a short delivery time. Direct contacts between the sponsor and our staff result in efficient scientific and technical support. We provide free consultation on next steps in research if desired by the sponsor.
 Quality. Flexibility. Short Turnaround Time.
Quality. Flexibility. Short Turnaround Time.
References
Grobben et al. (2020) Structural insights into human Arginase-1 pH dependence and its inhibition by the small molecule inhibitor CB-1158. Journal of Structural Biology: X, 4:100014.
Grobben et al. (2020) High-throughput fluorescence-based activity assay for Arginase-1. SLAS Discovery, 25 (9):1018-1025.
Uitdehaag et al. (2017) Target Residence Time-Guided Optimization on TTK Kinase Results in Inhibitors with Potent Anti-Proliferative Activity. Journal of Molecular Biology, 429:2211-2230.